Weighted Averages Scores for Species
wascores.RdComputes Weighted Averages scores of species for ordination configuration or for environmental variables.
Arguments
- x
Environmental variables or ordination scores, or for
wascoresobject withstdev = TRUE.- w
Weights: species abundances.
- expand
Expand weighted averages so that they have the same weighted variance as the corresponding environmental variables.
- stdev
Estimate weighted standard deviation of WA scores.
- display
Type of scores returned.
- ...
Other arguments passed to functions (currently ignored).
Details
Weighted Averages are a classical way of estimating the species
optima along continuous environmental variables
(a.k.a. gradients). Function wascores is a simple function
that is mainly designed to add species scores to unimodal
ordinations (metaMDS, sppscores) or
ordering rows or columns to give diagonal pattern of tabulation
(vegemite, tabasco). It can also be used
to find species “optima” or sampling unit calibrations for
community data. For this purpose, specialized packages such
analogue are recommended (but see
calibrate.cca).
First argument of wascores is the variable or a matrix of
variables for which weighted averages are needed, and the second
argument is the matrix of weights. In classical approaches weights are
a community matrix, where taxon abundances define the weights. The
number of rows must match. If the first argument is for taxa
(columns), community weight matrix must be transposed.
Weighted averages “shrink”: they cannot be more extreme than
values used for calculating the averages. With expand = TRUE,
the function “deshrinks” the weighted averages making their
weighted variance equal to the weighted variance of the
corresponding input variable. Specialized packages (such as
analogue) offer a wider range of deshrinking alternatives, but
deshrinking can also made after the analysis (see
Examples). Function eigengrad returns the strength of
expansion as attribute shrinkage of the wascores
result for each environmental gradient. The shrinkage equal to the
constrained eigenvalue of cca when only this one
gradient was used as a constraint, and describes the strength of the
gradient.
With stdev = TRUE the function estimates the unbiased
weighted standard deviation of the WA estimates using
cov.wt. For unbiased standard deviation the virtual
number of observations is equal to inverse Simpson index of
diversity also known as Hill number N2 (see
diversity). The numeric results can be accessed with
scores function. Function tolerance uses the
same algebra for weighted standard deviation, but bases the variance
on linear combination scores (constaints) variables instead of the
weighted averages of the sites like wascores.
Weighted averages are closely linked to correspondence analysis
(ca, cca). Repeated use of
wascores will converge to the first axis of unconstrained
correspondence analysis (ca) which therefore is also
known as Reciprocal Averaging (Hill 1973). Constrained
correspondence analysis (cca) is equivalent to
weighted averages and calibrate.cca will return
weighted averages of the constraint with different deshrinking.
Value
If stdev = TRUE, function returns an object of class
"wascores" with items
- wa
A matrix of weighted averages with. If
expand=TRUE, attributeshrinkagehas the inverses of squared expansion factors orccaeigenvalues for the variable and attributecentrefor the weighted means of the variables.- stdev
a matrix of weighted standard deviations
- n2
effective sample sizes which are equal to inverse Simpson diversity or Hill number N2
If stdev = FALSE (default), only the plain matrix wa
is returned. Function eigengrad returns only the
shrinkage attribute. With stdev = TRUE only a brief
summary of the result is printed, and the individvual scores can be
accessed with scores function.
References
Hill, M.O. (1973) Reciprocal averaging: An eigenvector method of ordination. Journal of Ecology 61, 237–249.
Examples
data(mite, mite.env)
## add species points to ordination
mod <- monoMDS(vegdist(mite))
plot(mod)
## add species points; sppscores does the same and can also add the
## species scores to mod
points(wascores(scores(mod), mite, expand = TRUE), pch="+", col=2)
 ## Get taxon optima for WatrCont
head(wascores(mite.env$WatrCont, mite))
#> [,1]
#> Brachy 360.4302
#> PHTH 292.0329
#> HPAV 392.4000
#> RARD 277.4195
#> SSTR 359.1609
#> Protopl 248.4969
## WA calibration: site WA from species WA; NB using transpose for site WA
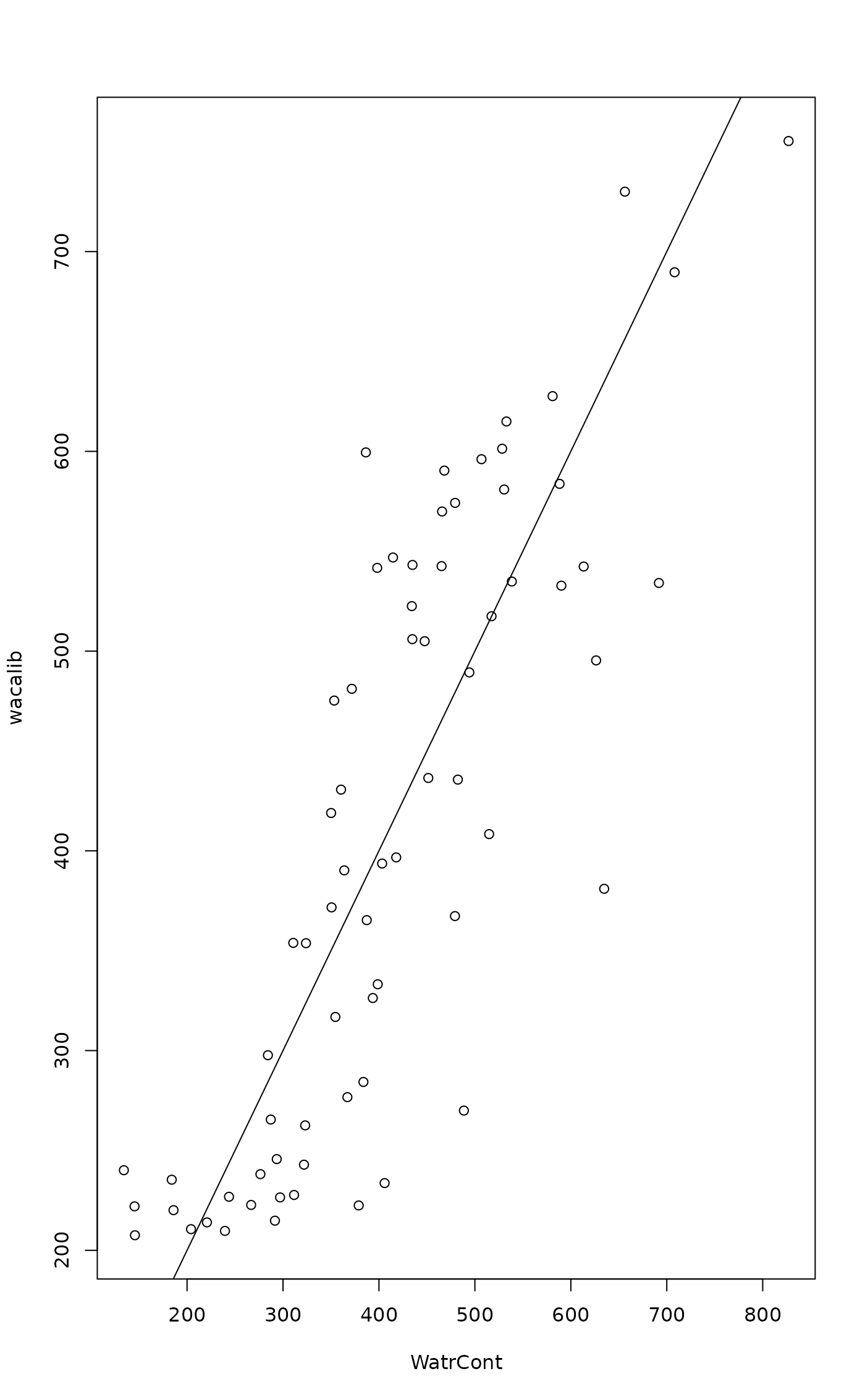
spwa <- wascores(mite.env$WatrCont, mite, expand = TRUE)
wacalib <- wascores(spwa, t(mite), expand = TRUE)
plot(wacalib ~ WatrCont, data=mite.env)
abline(0, 1)
## Get taxon optima for WatrCont
head(wascores(mite.env$WatrCont, mite))
#> [,1]
#> Brachy 360.4302
#> PHTH 292.0329
#> HPAV 392.4000
#> RARD 277.4195
#> SSTR 359.1609
#> Protopl 248.4969
## WA calibration: site WA from species WA; NB using transpose for site WA
spwa <- wascores(mite.env$WatrCont, mite, expand = TRUE)
wacalib <- wascores(spwa, t(mite), expand = TRUE)
plot(wacalib ~ WatrCont, data=mite.env)
abline(0, 1)
 ## use traditional 'inverse' regression deshrinking instead of wascores
## 'expand'
wareg <- fitted(lm(WatrCont ~ wacalib, data=mite.env))
head(cbind("WatrCont" = mite.env$WatrCont, "expand" = drop(wacalib),
"regression" = wareg))
#> WatrCont expand regression
#> 1 350.15 418.9468 418.3084
#> 2 434.81 505.9779 484.6130
#> 3 371.72 481.1096 465.6672
#> 4 360.50 430.6437 427.2198
#> 5 204.13 210.6019 259.5811
#> 6 311.55 227.7218 272.6239
## Reciprocal Averaging algorithm for Correspondence Analysis
## start with random values
u <- runif(nrow(mite))
## repeat the following steps so long that the shrinkage converges
v <- wascores(u, mite, expand = TRUE)
u <- wascores(v, t(mite), expand = TRUE)
attr(u, "shrinkage") # current estimate of eigenvalue
#> [1] 0.037767
## The strengths of two continuous variables in the data set
eigengrad(mite.env[, 1:2], mite)
#> SubsDens WatrCont
#> 0.09996798 0.39512786
## use traditional 'inverse' regression deshrinking instead of wascores
## 'expand'
wareg <- fitted(lm(WatrCont ~ wacalib, data=mite.env))
head(cbind("WatrCont" = mite.env$WatrCont, "expand" = drop(wacalib),
"regression" = wareg))
#> WatrCont expand regression
#> 1 350.15 418.9468 418.3084
#> 2 434.81 505.9779 484.6130
#> 3 371.72 481.1096 465.6672
#> 4 360.50 430.6437 427.2198
#> 5 204.13 210.6019 259.5811
#> 6 311.55 227.7218 272.6239
## Reciprocal Averaging algorithm for Correspondence Analysis
## start with random values
u <- runif(nrow(mite))
## repeat the following steps so long that the shrinkage converges
v <- wascores(u, mite, expand = TRUE)
u <- wascores(v, t(mite), expand = TRUE)
attr(u, "shrinkage") # current estimate of eigenvalue
#> [1] 0.037767
## The strengths of two continuous variables in the data set
eigengrad(mite.env[, 1:2], mite)
#> SubsDens WatrCont
#> 0.09996798 0.39512786