Diagnostic Tools for [Constrained] Ordination (CCA, RDA, DCA, CA, PCA)
goodness.cca.RdFunctions goodness and inertcomp can
be used to assess the goodness of fit for individual sites or
species. Function vif.cca and alias.cca can be used to
analyse linear dependencies among constraints and conditions. In
addition, there are some other diagnostic tools (see 'Details').
Usage
# S3 method for class 'cca'
goodness(object, choices, display = c("species", "sites"),
model = c("CCA", "CA"), summarize = FALSE, addprevious = FALSE, ...)
inertcomp(object, display = c("species", "sites"),
unity = FALSE, proportional = FALSE)
spenvcor(object)
intersetcor(object)
vif.cca(object)
# S3 method for class 'cca'
alias(object, names.only = FALSE, ...)Arguments
- object
- display
Display
"species"or"sites". Species are not available indbrdaandcapscale.- choices
Axes shown. Default is to show all axes of the
"model".- model
Show constrained (
"CCA") or unconstrained ("CA") results.- summarize
Show only the accumulated total.
- addprevious
Add the variation explained by previous components when
statistic="explained". Formodel = "CCA"add conditioned (partialled out) variation, and formodel = "CA"add both conditioned and constrained variation. This will give cumulative explanation with previous components.- unity
Scale inertia components to unit sum (sum of all items is 1).
- proportional
Give the inertia components as proportional for the corresponding total of the item (sum of each row is 1). This option takes precedence over
unity.- names.only
Return only names of aliased variable(s) instead of defining equations.
- ...
Other parameters to the functions.
Details
Function goodness gives cumulative proportion of inertia
accounted by species up to chosen axes. The proportions can be
assessed either by species or by sites depending on the argument
display, but species are not available in distance-based
dbrda. The function is not implemented for
capscale.
Function inertcomp decomposes the inertia into partial,
constrained and unconstrained components for each site or species.
Legendre & De Cáceres (2012) called these inertia
components as local contributions to beta-diversity (LCBD) and
species contributions to beta-diversity (SCBD), and they give these
as relative contributions summing up to unity (argument
unity = TRUE). For this interpretation, appropriate dissimilarity
measures should be used in dbrda or appropriate
standardization in rda (Legendre & De
Cáceres 2012). The function is not implemented for
capscale.
Function spenvcor finds the so-called “species –
environment correlation” or (weighted) correlation of
weighted average scores and linear combination scores. This is a bad
measure of goodness of ordination, because it is sensitive to extreme
scores (like correlations are), and very sensitive to overfitting or
using too many constraints. Better models often have poorer
correlations. Function ordispider can show the same
graphically.
Function intersetcor finds the so-called “interset
correlation” or (weighted) correlation of weighted averages scores
and constraints. The defined contrasts are used for factor
variables. This is a bad measure since it is a correlation. Further,
it focuses on correlations between single contrasts and single axes
instead of looking at the multivariate relationship. Fitted vectors
(envfit) provide a better alternative. Biplot scores
(see scores.cca) are a multivariate alternative for
(weighted) correlation between linear combination scores and
constraints.
Function vif.cca gives the variance inflation factors for each
constraint or contrast in factor constraints. In partial ordination,
conditioning variables are analysed together with constraints. Variance
inflation is a diagnostic tool to identify useless constraints. A
common rule is that values over 10 indicate redundant
constraints. If later constraints are complete linear combinations of
conditions or previous constraints, they will be completely removed
from the estimation, and no biplot scores or centroids are calculated
for these aliased constraints. A note will be printed with default
output if there are aliased constraints. Function alias will
give the linear coefficients defining the aliased constraints, or
only their names with argument names.only = TRUE.
References
Greenacre, M. J. (1984). Theory and applications of correspondence analysis. Academic Press, London.
Gross, J. (2003). Variance inflation factors. R News 3(1), 13–15.
Legendre, P. & De Cáceres, M. (2012). Beta diversity as the variance of community data: dissimilarity coefficients and partitioning. Ecology Letters 16, 951–963. doi:10.1111/ele.12141
Author
Jari Oksanen. The vif.cca relies heavily on the code by
W. N. Venables. alias.cca is a simplified version of
alias.lm.
Examples
data(dune, dune.env)
mod <- cca(dune ~ A1 + Management + Condition(Moisture), data=dune.env)
goodness(mod, addprevious = TRUE)
#> CCA1 CCA2 CCA3 CCA4
#> Achimill 0.36630013 0.3822685 0.3838616 0.4934158
#> Agrostol 0.67247051 0.6724758 0.6779597 0.7773267
#> Airaprae 0.36213737 0.3698100 0.3816619 0.3908018
#> Alopgeni 0.61547145 0.6966105 0.7042650 0.7212918
#> Anthodor 0.24619147 0.2795001 0.3509172 0.3609709
#> Bellpere 0.41185412 0.4179432 0.4847618 0.4849622
#> Bromhord 0.33487622 0.3397416 0.3870032 0.5505037
#> Chenalbu 0.23594716 0.2684323 0.2828928 0.2885321
#> Cirsarve 0.29041563 0.3013655 0.3080671 0.3591280
#> Comapalu 0.16338257 0.6836790 0.7390659 0.7963425
#> Eleopalu 0.55132024 0.6099415 0.6193301 0.6259818
#> Elymrepe 0.25239595 0.2710266 0.2761491 0.2882666
#> Empenigr 0.27089495 0.3132399 0.3153052 0.3154203
#> Hyporadi 0.31349648 0.3371809 0.3387669 0.3388716
#> Juncarti 0.43923609 0.4492937 0.4871043 0.5224072
#> Juncbufo 0.70439967 0.7226263 0.7228786 0.7257471
#> Lolipere 0.48141171 0.5720410 0.5727299 0.6034007
#> Planlanc 0.54969676 0.6084389 0.6802195 0.6826265
#> Poaprat 0.40267189 0.4944813 0.5014516 0.5326546
#> Poatriv 0.49694972 0.5409439 0.5468830 0.5594817
#> Ranuflam 0.68677962 0.6983001 0.7020461 0.7064850
#> Rumeacet 0.44788204 0.5211145 0.7673956 0.7691199
#> Sagiproc 0.27039747 0.3497634 0.3553109 0.3613746
#> Salirepe 0.64788354 0.7264891 0.7276110 0.7639711
#> Scorautu 0.54312496 0.5510319 0.6078931 0.6140593
#> Trifprat 0.37328840 0.4101104 0.6624199 0.6625703
#> Trifrepe 0.03048149 0.2115857 0.3300132 0.4207437
#> Vicilath 0.17824132 0.1784611 0.3762406 0.4279428
#> Bracruta 0.15585567 0.1641095 0.1672797 0.2449864
#> Callcusp 0.30771429 0.3143582 0.3308502 0.3518027
goodness(mod, addprevious = TRUE, summ = TRUE)
#> Achimill Agrostol Airaprae Alopgeni Anthodor Bellpere Bromhord Chenalbu
#> 0.4934158 0.7773267 0.3908018 0.7212918 0.3609709 0.4849622 0.5505037 0.2885321
#> Cirsarve Comapalu Eleopalu Elymrepe Empenigr Hyporadi Juncarti Juncbufo
#> 0.3591280 0.7963425 0.6259818 0.2882666 0.3154203 0.3388716 0.5224072 0.7257471
#> Lolipere Planlanc Poaprat Poatriv Ranuflam Rumeacet Sagiproc Salirepe
#> 0.6034007 0.6826265 0.5326546 0.5594817 0.7064850 0.7691199 0.3613746 0.7639711
#> Scorautu Trifprat Trifrepe Vicilath Bracruta Callcusp
#> 0.6140593 0.6625703 0.4207437 0.4279428 0.2449864 0.3518027
## Fit of species in 2 dimensions
good <- goodness(mod, choices = 1:2, summarize = TRUE)
sort(good)
#> Bellpere Bromhord Ranuflam Juncarti Callcusp Cirsarve
#> 0.008218774 0.009274805 0.013201031 0.014901783 0.019884022 0.034866365
#> Achimill Elymrepe Juncbufo Vicilath Trifprat Eleopalu
#> 0.039549538 0.048683379 0.056399541 0.057206719 0.064662265 0.070393304
#> Planlanc Sagiproc Bracruta Poaprat Lolipere Agrostol
#> 0.088501412 0.089259090 0.091882493 0.100400726 0.109310553 0.116451764
#> Rumeacet Chenalbu Anthodor Trifrepe Empenigr Hyporadi
#> 0.119854640 0.157788824 0.176908672 0.190263830 0.209619927 0.298284611
#> Airaprae Alopgeni Scorautu Poatriv Comapalu Salirepe
#> 0.305762791 0.354220783 0.442077524 0.484960397 0.522716229 0.601210690
## Drop poorly fitting species from a plot
plot(mod, spe.par = list(select = good > 0.1, optimize = TRUE,
bg = "yellow"))
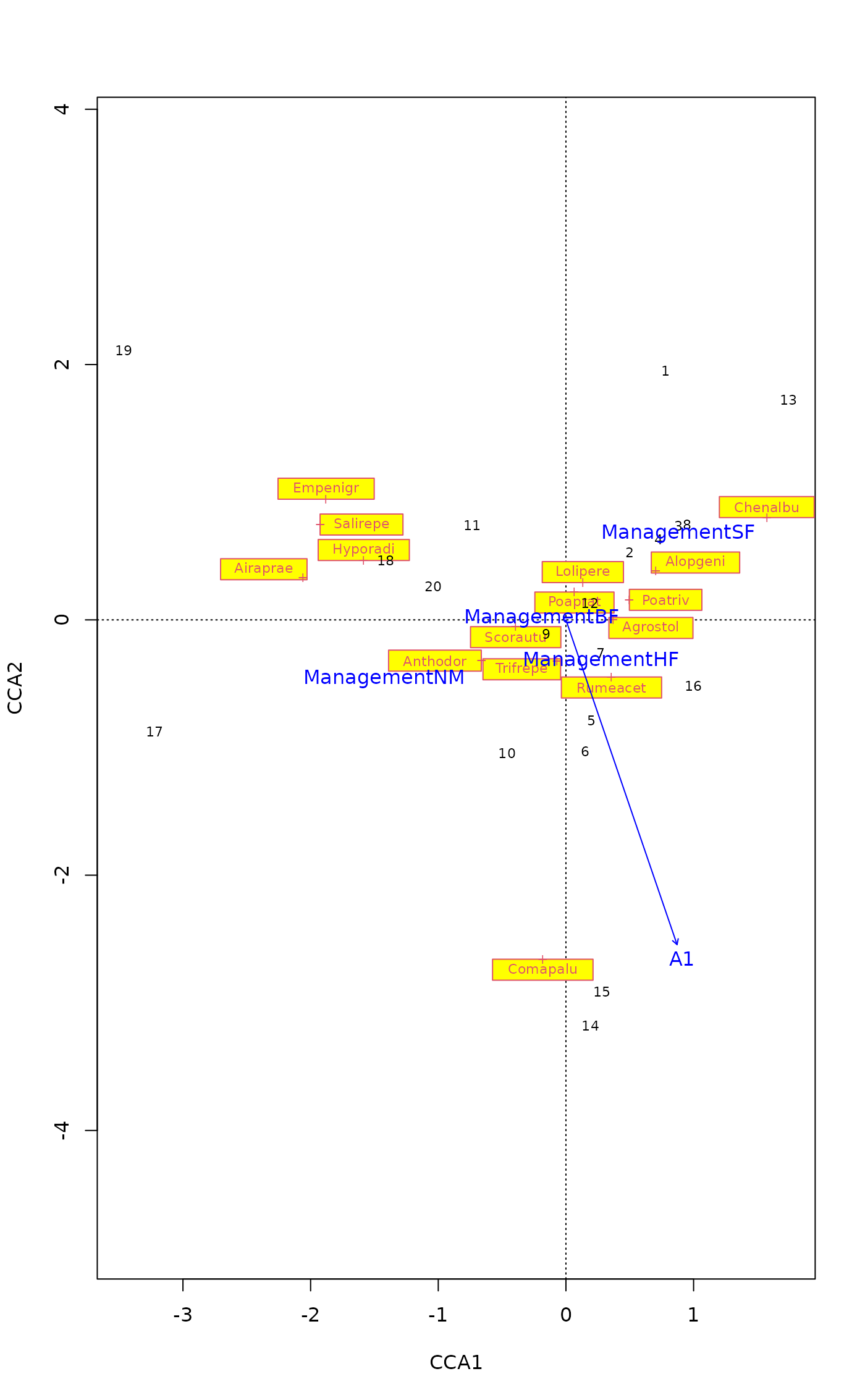 ## Inertia components
inertcomp(mod, prop = TRUE)
#> pCCA CCA CA
#> Achimill 0.34271900 0.15069678 0.5065842
#> Agrostol 0.55602406 0.22130269 0.2226733
#> Airaprae 0.06404726 0.32675457 0.6091982
#> Alopgeni 0.34238968 0.37890210 0.2787082
#> Anthodor 0.10259139 0.25837947 0.6390291
#> Bellpere 0.40972447 0.07523776 0.5150378
#> Bromhord 0.33046684 0.22003683 0.4494963
#> Chenalbu 0.11064346 0.17788865 0.7114679
#> Cirsarve 0.26649913 0.09262886 0.6408720
#> Comapalu 0.16096277 0.63537969 0.2036575
#> Eleopalu 0.53954819 0.08643366 0.3740182
#> Elymrepe 0.22234322 0.06592337 0.7117334
#> Empenigr 0.10361994 0.21180040 0.6845797
#> Hyporadi 0.03889627 0.29997533 0.6611284
#> Juncarti 0.43439190 0.08801527 0.4775928
#> Juncbufo 0.66622672 0.05952038 0.2742529
#> Lolipere 0.46273045 0.14067027 0.3965993
#> Planlanc 0.51993753 0.16268893 0.3173735
#> Poaprat 0.39408053 0.13857406 0.4673454
#> Poatriv 0.05598349 0.50349824 0.4405183
#> Ranuflam 0.68509904 0.02138594 0.2935150
#> Rumeacet 0.40125987 0.36786003 0.2308801
#> Sagiproc 0.26050435 0.10087025 0.6386254
#> Salirepe 0.12527838 0.63869277 0.2360289
#> Scorautu 0.10895437 0.50510492 0.3859407
#> Trifprat 0.34544815 0.31712212 0.3374297
#> Trifrepe 0.02132183 0.39942191 0.5792563
#> Vicilath 0.12125433 0.30668844 0.5720572
#> Bracruta 0.07222706 0.17275938 0.7550136
#> Callcusp 0.29447422 0.05732850 0.6481973
inertcomp(mod)
#> pCCA CCA CA
#> Achimill 0.0173766015 0.007640656 0.02568493
#> Agrostol 0.0456558521 0.018171449 0.01828399
#> Airaprae 0.0066672285 0.034014687 0.06341666
#> Alopgeni 0.0325977567 0.036073980 0.02653486
#> Anthodor 0.0096274015 0.024246897 0.05996790
#> Bellpere 0.0154640710 0.002839669 0.01943887
#> Bromhord 0.0180126793 0.011993496 0.02450059
#> Chenalbu 0.0031913088 0.005130874 0.02052099
#> Cirsarve 0.0110663060 0.003846389 0.02661204
#> Comapalu 0.0127652351 0.050389111 0.01615116
#> Eleopalu 0.0797827194 0.012780901 0.05530588
#> Elymrepe 0.0193932154 0.005749967 0.06207879
#> Empenigr 0.0063826176 0.013046147 0.04216766
#> Hyporadi 0.0046669914 0.035992710 0.07932587
#> Juncarti 0.0359126341 0.007276518 0.03948420
#> Juncbufo 0.0494087668 0.004414156 0.02033917
#> Lolipere 0.0368344271 0.011197683 0.03157023
#> Planlanc 0.0366139947 0.011456552 0.02234944
#> Poaprat 0.0142991623 0.005028142 0.01695757
#> Poatriv 0.0028845344 0.025942611 0.02269759
#> Ranuflam 0.0446783229 0.001394671 0.01914141
#> Rumeacet 0.0288221948 0.026423110 0.01658394
#> Sagiproc 0.0151161507 0.005853146 0.03705718
#> Salirepe 0.0142756439 0.072779924 0.02689581
#> Scorautu 0.0030643984 0.014206339 0.01085478
#> Trifprat 0.0228613139 0.020986733 0.02233067
#> Trifrepe 0.0008339368 0.015622139 0.02265580
#> Vicilath 0.0049088357 0.012415912 0.02315905
#> Bracruta 0.0032317812 0.007730074 0.03378289
#> Callcusp 0.0319130878 0.006212868 0.07024716
## vif.cca
vif.cca(mod)
#> Moisture.L Moisture.Q Moisture.C A1 ManagementHF ManagementNM
#> 1.504327 1.284489 1.347660 1.367328 2.238653 2.570972
#> ManagementSF
#> 2.424444
## Aliased constraints
mod <- cca(dune ~ ., dune.env)
#>
#> Some constraints or conditions were aliased because they were redundant. This
#> can happen if terms are constant or linearly dependent (collinear): ‘Manure^4’
mod
#>
#> Call: cca(formula = dune ~ A1 + Moisture + Management + Use + Manure, data
#> = dune.env)
#>
#> Inertia Proportion Rank
#> Total 2.1153 1.0000
#> Constrained 1.5032 0.7106 12
#> Unconstrained 0.6121 0.2894 7
#>
#> Inertia is scaled Chi-square
#>
#> -- NOTE:
#> Some constraints or conditions were aliased because they were redundant.
#> This can happen if terms are constant or linearly dependent (collinear):
#> ‘Manure^4’
#>
#> Eigenvalues for constrained axes:
#> CCA1 CCA2 CCA3 CCA4 CCA5 CCA6 CCA7 CCA8 CCA9 CCA10 CCA11
#> 0.4671 0.3410 0.1761 0.1532 0.0953 0.0703 0.0589 0.0499 0.0318 0.0260 0.0228
#> CCA12
#> 0.0108
#>
#> Eigenvalues for unconstrained axes:
#> CA1 CA2 CA3 CA4 CA5 CA6 CA7
#> 0.27237 0.10876 0.08975 0.06305 0.03489 0.02529 0.01798
#>
vif.cca(mod)
#> A1 Moisture.L Moisture.Q Moisture.C ManagementHF ManagementNM
#> 2.208249 2.858927 3.072715 3.587087 6.608315 142.359372
#> ManagementSF Use.L Use.Q Manure.L Manure.Q Manure.C
#> 12.862713 2.642718 3.007238 80.828330 49.294455 21.433337
#> Manure^4
#> NA
alias(mod)
#> Model :
#> dune ~ A1 + Moisture + Management + Use + Manure
#>
#> Complete :
#> A1 Moisture.L Moisture.Q Moisture.C ManagementHF ManagementNM
#> Manure^4 8.366600
#> ManagementSF Use.L Use.Q Manure.L Manure.Q Manure.C
#> Manure^4 5.291503 -4.472136 2.645751
#>
with(dune.env, table(Management, Manure))
#> Manure
#> Management 0 1 2 3 4
#> BF 0 2 1 0 0
#> HF 0 1 2 2 0
#> NM 6 0 0 0 0
#> SF 0 0 1 2 3
## The standard correlations (not recommended)
## IGNORE_RDIFF_BEGIN
spenvcor(mod)
#> CCA1 CCA2 CCA3 CCA4 CCA5 CCA6 CCA7 CCA8
#> 0.9636709 0.9487249 0.9330741 0.8734876 0.9373716 0.8362687 0.9748793 0.8392720
#> CCA9 CCA10 CCA11 CCA12
#> 0.8748741 0.6087512 0.6633248 0.7581210
intersetcor(mod)
#> CCA1 CCA2 CCA3 CCA4 CCA5
#> A1 -0.5332506 0.13691202 -0.47996401 -0.259859587 -0.09894964
#> Moisture.L -0.8785505 0.17867589 0.03714134 0.181952935 -0.09826534
#> Moisture.Q -0.1956664 -0.33044917 -0.27321286 -0.180333890 0.26609291
#> Moisture.C -0.2023782 -0.09698397 0.28596824 -0.261712720 -0.49103002
#> ManagementHF 0.3473460 0.01680324 -0.51205769 0.194144965 0.30752664
#> ManagementNM -0.5699549 -0.61111645 0.14751127 -0.013777789 0.04571982
#> ManagementSF -0.1197499 0.64084416 0.19780650 0.134892908 -0.09679992
#> Use.L -0.1871999 0.32990444 -0.30941161 -0.372747011 0.09586963
#> Use.Q -0.1820298 -0.48874152 -0.01997442 -0.009812946 0.04812588
#> Manure.L 0.3175126 0.65945634 0.03724864 -0.025383543 -0.04077470
#> Manure.Q -0.4075615 -0.21149073 0.49297244 -0.176686201 0.11973190
#> Manure.C 0.4676279 0.11376054 0.29132473 -0.173382982 0.14219924
#> Manure^4 0.2222349 -0.12789494 -0.12921227 0.108367170 -0.02559567
#> CCA6 CCA7 CCA8 CCA9 CCA10
#> A1 -0.15225816 0.25788462 0.19247720 -0.27694466 -0.1158449480
#> Moisture.L -0.02923342 0.07858647 -0.10772510 0.07101300 0.0952517164
#> Moisture.Q -0.11211675 0.05062810 -0.48302647 0.06138704 -0.2053304965
#> Moisture.C -0.23581275 -0.38693407 -0.10144580 -0.21907160 0.1875632770
#> ManagementHF -0.24278705 0.16364055 -0.14053438 0.31066725 0.1310215145
#> ManagementNM -0.06430101 0.23917584 0.14375754 -0.27103732 0.0002768613
#> ManagementSF -0.01611984 -0.49726250 0.08073472 -0.30235728 -0.1381281272
#> Use.L 0.19127262 -0.44624831 -0.18450714 0.12950951 0.0452826749
#> Use.Q 0.13485545 0.10367354 -0.11020112 0.41245485 -0.0766932005
#> Manure.L -0.22265819 -0.49627772 -0.16971786 -0.03943343 -0.0045229147
#> Manure.Q -0.19402211 -0.11937394 0.17611673 -0.44002593 0.0903998202
#> Manure.C 0.14760330 0.07842345 0.37774417 0.10181374 0.1055057288
#> Manure^4 -0.36683782 0.05953330 0.40927409 -0.06054381 -0.1500198368
#> CCA11 CCA12
#> A1 -0.03550223 -0.08881387
#> Moisture.L 0.06404776 -0.08587882
#> Moisture.Q -0.21810558 0.16917878
#> Moisture.C 0.13701079 -0.14260914
#> ManagementHF 0.17283125 0.13296499
#> ManagementNM -0.01358436 0.09533598
#> ManagementSF -0.01468592 -0.06614834
#> Use.L -0.08584883 0.32559307
#> Use.Q 0.41893616 0.04881247
#> Manure.L 0.02396993 0.13049087
#> Manure.Q 0.12987366 0.07137031
#> Manure.C 0.05176927 -0.41550238
#> Manure^4 -0.41603287 0.01661279
## IGNORE_RDIFF_END
## Inertia components
inertcomp(mod, prop = TRUE)
#> pCCA CCA CA
#> Achimill 0.34271900 0.15069678 0.5065842
#> Agrostol 0.55602406 0.22130269 0.2226733
#> Airaprae 0.06404726 0.32675457 0.6091982
#> Alopgeni 0.34238968 0.37890210 0.2787082
#> Anthodor 0.10259139 0.25837947 0.6390291
#> Bellpere 0.40972447 0.07523776 0.5150378
#> Bromhord 0.33046684 0.22003683 0.4494963
#> Chenalbu 0.11064346 0.17788865 0.7114679
#> Cirsarve 0.26649913 0.09262886 0.6408720
#> Comapalu 0.16096277 0.63537969 0.2036575
#> Eleopalu 0.53954819 0.08643366 0.3740182
#> Elymrepe 0.22234322 0.06592337 0.7117334
#> Empenigr 0.10361994 0.21180040 0.6845797
#> Hyporadi 0.03889627 0.29997533 0.6611284
#> Juncarti 0.43439190 0.08801527 0.4775928
#> Juncbufo 0.66622672 0.05952038 0.2742529
#> Lolipere 0.46273045 0.14067027 0.3965993
#> Planlanc 0.51993753 0.16268893 0.3173735
#> Poaprat 0.39408053 0.13857406 0.4673454
#> Poatriv 0.05598349 0.50349824 0.4405183
#> Ranuflam 0.68509904 0.02138594 0.2935150
#> Rumeacet 0.40125987 0.36786003 0.2308801
#> Sagiproc 0.26050435 0.10087025 0.6386254
#> Salirepe 0.12527838 0.63869277 0.2360289
#> Scorautu 0.10895437 0.50510492 0.3859407
#> Trifprat 0.34544815 0.31712212 0.3374297
#> Trifrepe 0.02132183 0.39942191 0.5792563
#> Vicilath 0.12125433 0.30668844 0.5720572
#> Bracruta 0.07222706 0.17275938 0.7550136
#> Callcusp 0.29447422 0.05732850 0.6481973
inertcomp(mod)
#> pCCA CCA CA
#> Achimill 0.0173766015 0.007640656 0.02568493
#> Agrostol 0.0456558521 0.018171449 0.01828399
#> Airaprae 0.0066672285 0.034014687 0.06341666
#> Alopgeni 0.0325977567 0.036073980 0.02653486
#> Anthodor 0.0096274015 0.024246897 0.05996790
#> Bellpere 0.0154640710 0.002839669 0.01943887
#> Bromhord 0.0180126793 0.011993496 0.02450059
#> Chenalbu 0.0031913088 0.005130874 0.02052099
#> Cirsarve 0.0110663060 0.003846389 0.02661204
#> Comapalu 0.0127652351 0.050389111 0.01615116
#> Eleopalu 0.0797827194 0.012780901 0.05530588
#> Elymrepe 0.0193932154 0.005749967 0.06207879
#> Empenigr 0.0063826176 0.013046147 0.04216766
#> Hyporadi 0.0046669914 0.035992710 0.07932587
#> Juncarti 0.0359126341 0.007276518 0.03948420
#> Juncbufo 0.0494087668 0.004414156 0.02033917
#> Lolipere 0.0368344271 0.011197683 0.03157023
#> Planlanc 0.0366139947 0.011456552 0.02234944
#> Poaprat 0.0142991623 0.005028142 0.01695757
#> Poatriv 0.0028845344 0.025942611 0.02269759
#> Ranuflam 0.0446783229 0.001394671 0.01914141
#> Rumeacet 0.0288221948 0.026423110 0.01658394
#> Sagiproc 0.0151161507 0.005853146 0.03705718
#> Salirepe 0.0142756439 0.072779924 0.02689581
#> Scorautu 0.0030643984 0.014206339 0.01085478
#> Trifprat 0.0228613139 0.020986733 0.02233067
#> Trifrepe 0.0008339368 0.015622139 0.02265580
#> Vicilath 0.0049088357 0.012415912 0.02315905
#> Bracruta 0.0032317812 0.007730074 0.03378289
#> Callcusp 0.0319130878 0.006212868 0.07024716
## vif.cca
vif.cca(mod)
#> Moisture.L Moisture.Q Moisture.C A1 ManagementHF ManagementNM
#> 1.504327 1.284489 1.347660 1.367328 2.238653 2.570972
#> ManagementSF
#> 2.424444
## Aliased constraints
mod <- cca(dune ~ ., dune.env)
#>
#> Some constraints or conditions were aliased because they were redundant. This
#> can happen if terms are constant or linearly dependent (collinear): ‘Manure^4’
mod
#>
#> Call: cca(formula = dune ~ A1 + Moisture + Management + Use + Manure, data
#> = dune.env)
#>
#> Inertia Proportion Rank
#> Total 2.1153 1.0000
#> Constrained 1.5032 0.7106 12
#> Unconstrained 0.6121 0.2894 7
#>
#> Inertia is scaled Chi-square
#>
#> -- NOTE:
#> Some constraints or conditions were aliased because they were redundant.
#> This can happen if terms are constant or linearly dependent (collinear):
#> ‘Manure^4’
#>
#> Eigenvalues for constrained axes:
#> CCA1 CCA2 CCA3 CCA4 CCA5 CCA6 CCA7 CCA8 CCA9 CCA10 CCA11
#> 0.4671 0.3410 0.1761 0.1532 0.0953 0.0703 0.0589 0.0499 0.0318 0.0260 0.0228
#> CCA12
#> 0.0108
#>
#> Eigenvalues for unconstrained axes:
#> CA1 CA2 CA3 CA4 CA5 CA6 CA7
#> 0.27237 0.10876 0.08975 0.06305 0.03489 0.02529 0.01798
#>
vif.cca(mod)
#> A1 Moisture.L Moisture.Q Moisture.C ManagementHF ManagementNM
#> 2.208249 2.858927 3.072715 3.587087 6.608315 142.359372
#> ManagementSF Use.L Use.Q Manure.L Manure.Q Manure.C
#> 12.862713 2.642718 3.007238 80.828330 49.294455 21.433337
#> Manure^4
#> NA
alias(mod)
#> Model :
#> dune ~ A1 + Moisture + Management + Use + Manure
#>
#> Complete :
#> A1 Moisture.L Moisture.Q Moisture.C ManagementHF ManagementNM
#> Manure^4 8.366600
#> ManagementSF Use.L Use.Q Manure.L Manure.Q Manure.C
#> Manure^4 5.291503 -4.472136 2.645751
#>
with(dune.env, table(Management, Manure))
#> Manure
#> Management 0 1 2 3 4
#> BF 0 2 1 0 0
#> HF 0 1 2 2 0
#> NM 6 0 0 0 0
#> SF 0 0 1 2 3
## The standard correlations (not recommended)
## IGNORE_RDIFF_BEGIN
spenvcor(mod)
#> CCA1 CCA2 CCA3 CCA4 CCA5 CCA6 CCA7 CCA8
#> 0.9636709 0.9487249 0.9330741 0.8734876 0.9373716 0.8362687 0.9748793 0.8392720
#> CCA9 CCA10 CCA11 CCA12
#> 0.8748741 0.6087512 0.6633248 0.7581210
intersetcor(mod)
#> CCA1 CCA2 CCA3 CCA4 CCA5
#> A1 -0.5332506 0.13691202 -0.47996401 -0.259859587 -0.09894964
#> Moisture.L -0.8785505 0.17867589 0.03714134 0.181952935 -0.09826534
#> Moisture.Q -0.1956664 -0.33044917 -0.27321286 -0.180333890 0.26609291
#> Moisture.C -0.2023782 -0.09698397 0.28596824 -0.261712720 -0.49103002
#> ManagementHF 0.3473460 0.01680324 -0.51205769 0.194144965 0.30752664
#> ManagementNM -0.5699549 -0.61111645 0.14751127 -0.013777789 0.04571982
#> ManagementSF -0.1197499 0.64084416 0.19780650 0.134892908 -0.09679992
#> Use.L -0.1871999 0.32990444 -0.30941161 -0.372747011 0.09586963
#> Use.Q -0.1820298 -0.48874152 -0.01997442 -0.009812946 0.04812588
#> Manure.L 0.3175126 0.65945634 0.03724864 -0.025383543 -0.04077470
#> Manure.Q -0.4075615 -0.21149073 0.49297244 -0.176686201 0.11973190
#> Manure.C 0.4676279 0.11376054 0.29132473 -0.173382982 0.14219924
#> Manure^4 0.2222349 -0.12789494 -0.12921227 0.108367170 -0.02559567
#> CCA6 CCA7 CCA8 CCA9 CCA10
#> A1 -0.15225816 0.25788462 0.19247720 -0.27694466 -0.1158449480
#> Moisture.L -0.02923342 0.07858647 -0.10772510 0.07101300 0.0952517164
#> Moisture.Q -0.11211675 0.05062810 -0.48302647 0.06138704 -0.2053304965
#> Moisture.C -0.23581275 -0.38693407 -0.10144580 -0.21907160 0.1875632770
#> ManagementHF -0.24278705 0.16364055 -0.14053438 0.31066725 0.1310215145
#> ManagementNM -0.06430101 0.23917584 0.14375754 -0.27103732 0.0002768613
#> ManagementSF -0.01611984 -0.49726250 0.08073472 -0.30235728 -0.1381281272
#> Use.L 0.19127262 -0.44624831 -0.18450714 0.12950951 0.0452826749
#> Use.Q 0.13485545 0.10367354 -0.11020112 0.41245485 -0.0766932005
#> Manure.L -0.22265819 -0.49627772 -0.16971786 -0.03943343 -0.0045229147
#> Manure.Q -0.19402211 -0.11937394 0.17611673 -0.44002593 0.0903998202
#> Manure.C 0.14760330 0.07842345 0.37774417 0.10181374 0.1055057288
#> Manure^4 -0.36683782 0.05953330 0.40927409 -0.06054381 -0.1500198368
#> CCA11 CCA12
#> A1 -0.03550223 -0.08881387
#> Moisture.L 0.06404776 -0.08587882
#> Moisture.Q -0.21810558 0.16917878
#> Moisture.C 0.13701079 -0.14260914
#> ManagementHF 0.17283125 0.13296499
#> ManagementNM -0.01358436 0.09533598
#> ManagementSF -0.01468592 -0.06614834
#> Use.L -0.08584883 0.32559307
#> Use.Q 0.41893616 0.04881247
#> Manure.L 0.02396993 0.13049087
#> Manure.Q 0.12987366 0.07137031
#> Manure.C 0.05176927 -0.41550238
#> Manure^4 -0.41603287 0.01661279
## IGNORE_RDIFF_END